May 07, 2013
By Matthew Chin
A research team led by UCLA bioengineers has developed a way to program and control the shape of fluids flowing through pipes or conduits without the need to solve complex and time-consuming fluid-motion equations.
This strategy, which the researchers liken to the major change that occurred when computer programmers no longer needed to understand the detailed physics behind computer circuits, could allow biologists, chemists, manufacturing engineers and others to tap the vast, unrealized potential of fluid-flow applications.
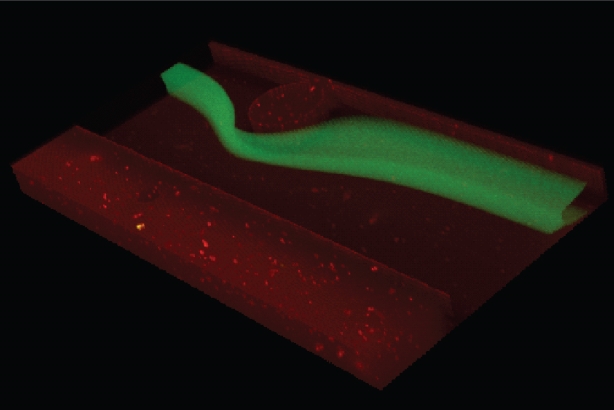
Co-flowing fluids are used today in a wide variety of applications. For example, medical diagnostic devices that analyze blood squeeze a central flow in a pipe with a surrounding flow — a process called hydrodynamic focusing — to achieve uniform measurements of cells within the blood. But the ability to control the shape of such fluid streams is limited and relies mostly on the relative rate at which two streams are introduced into the pipe.
The ability to more easily manipulate fluid streams into different shapes could help improve reactions in industrial plants, better cool computer chips, and shape and fabricate interlocking fibers to improve the properties of composite materials, the researchers said. Until now, however, such manipulations required a detailed knowledge of complex fluid mechanics and intricate numerical simulations.
The new strategy developed by the research team, which was led by Dino Di Carlo, associate professor of bioengineering at the UCLA Henry Samueli School of Engineering and Applied Science, eliminates the need to understand the mechanics of flow and makes designing new flows as simple as selecting books from the library.
The group successfully demonstrated a strategy in which the user picks from a library of many different fluid flow shapes. The shapes can be combined in series to create even more complex shapes. Once a user has settled on a final shape using these simple operations, the system provides the size and location of pillars sequenced within a channel that yield the shape, without the need for any fluid dynamic simulation. Thus, the expert mathematical knowledge of how to perform such a task is no longer required, and the design process is immensely sped up.
Just as computer programming underwent a revolution when the need to understand circuit physics was eliminated, this strategy could lead to new leaps in automation in biology, chemistry and materials science, the researchers said.
The paper is published in the peer-reviewed journal Nature Communications and is available online.
According to Di Carlo, the principal investigator on the research, such an approach opens up new ways of thinking about flows and how they can be used.
"Right now, using this technique, we are quickly designing simple fluid channels to perform automated operations on cells by moving and shaping fluid to wash cells and perform chemical dyeing procedures," Di Carlo said. "In the future, once we can tell a user how to design any shape of interest without trial and error, I see such an approach extending beyond the microscale to aid in shaping feed flows for manufacturing and chemical plants, maximizing light exposure to algae for biofuel production, or cooling large data-center warehouses efficiently."
"Importantly, the flow-sculpting approach we have developed can be used from very small to very large sizes — from channels smaller than a human hair filled with water to meter-scale pipes filled with oil — as long as certain parameters describing the flow remain constant," said lead author Hamed Amini, a former UCLA bioengineering doctoral student and postdoctoral scholar who is now a scientist at Illumina Inc.
The team also included Mahdokht Masaeli, a UCLA bioengineering graduate student; Elodie Sollier, a visiting scholar in Di Carlo's laboratory; Yu Xie, a mechanical engineering graduate student at Iowa State University; Baskar Ganapathysubramanian, an assistant professor of mechanical engineering at Iowa State University; and Howard A. Stone, the Donald R. Dixon '69 and Elizabeth W. Dixon Professor of Mechanical and Aerospace Engineering at Princeton University.
The research was funded by the National Science Foundation.
For more on Di Carlo's research, visit the Di Carlo Laboratory website.